This repository contains python code with datasets to run Raindrop algorithm. Raindrop is a graph-guided network for learning representations of irregularly sampled multivariate time series. We use Raindrop to classify time series of three healthcare and human activity datasets in four different settings.
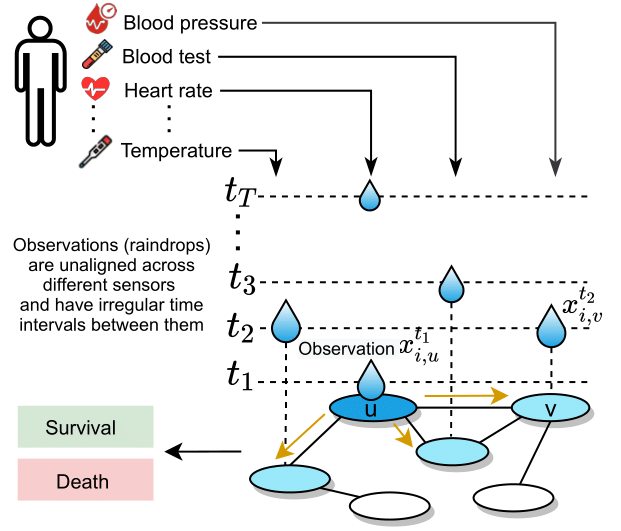
Raindrop is an approach, intended for learning representations of irregular multivariate time series, which models dependencies between sensors using neural message passing and temporal self attention. Raindrop represents every sample (e.g., patient) as a graph, where nodes indicate sensors and edges represent dependencies between them. Raindrop takes samples as input, each sample containing multiple sensors and each sensor consisting of irregularly recorded observations (e.g., in clinical data, an individual patient’s state of health, recorded at irregular time intervals with different subsets of sensors observed at different times). Raindrop model is inspired by the idea of raindrops falling into a pool at sequential but nonuniform time intervals and thereby creating ripple effects that propagate across the pool.
The main idea of Raindrop is to generate observation (a) and sensor (b) embeddings. Calculated sensor embeddings then serve as the basis for sample embeddings that can fed into a downstream task such as classification.
(a) Raindrop generates observation embedding based on observed value, passes message to neighbor sensors, and generates observation embedding through inter-sensor dependencies.
(b) An illustration of generating sensor embedding. We apply the message passing in (a) to all timestamps and produce corresponding observation embeddings. We aggregate arbitrary number of observation embeddings into a fixed-length sensor embedding, while paying distinctive attentions to different observations. We independently apply the processing procedure to all sensors.
We prepared to run our code for Raindrop as well as the baseline methods with two healthcare and one human activity dataset.
(1) P19 (PhysioNet Sepsis Early Prediction Challenge 2019) includes 38,803 patients that are monitored by 34 sensors. Each patient is associated with a binary label representing the occurrence of sepsis.
(2) P12 (PhysioNet Mortality Prediction Challenge 2012) records temporal measurements of 36 sensors of 11,988 patients in the first 48-hour stay in ICU. The samples are labeled based on hospitalization length.
(3) PAM (PAMAP2 Physical Activity Monitoring) contains 5,333 segments from 8 activities of daily living that are measured by 17 sensors.
The preprocessing scripts with data are available in folders P19data, P12data and PAMdata.
Let's look at the content of P12data folder, though, the structure is the same for all three folders with data. Some datasets may exclude raw data and do not have preprocessing scripts. Inside the *data folder, we have the following structure:
- process_scripts
- Inside we have preprocessing scripts and readme with the instructions how to run them.
- processed_data
- P_list.npy: Array of dictionaries, which is created from raw data. Array has a length of number of samples and each dictionary has keys 'id', 'static' variables and 'ts' time series data.
- PTdict_list.npy: Processed array of dictionaries. Array has a length of number of samples and each dictionary includes keys, such as 'id', 'static' attributes, 'arr' time series data and 'time' of observations.
- arr_outcomes.npy: The content has the shape (number of samples, outcomes). For each sample (patient) there are target outputs, such as length of hospital stay or mortality.
- ts_params.npy: Array with names of all sensors.
- static_params.npy: Array with names of static attributes.
- extended_static_params.npy: Array with names of extended static attributes (with more attributes than in static_params.npy).
- readme.md: Short description of the files.
- rawdata
- set-a: Data in the form of 4,000 .txt files, each containing time series observations.
- set-b: Data in the form of 4,000 .txt files, each containing time series observations.
- set-c: Data in the form of 4,000 .txt files, each containing time series observations.
- Outcomes-a: Text file, including target values (e.g., length of hospital stay, mortality) for all 4,000 samples from set-a.
- Outcomes-b: Text file, including target values (e.g., length of hospital stay, mortality) for all 4,000 samples from set-b.
- Outcomes-c: Text file, including target values (e.g., length of hospital stay, mortality) for all 4,000 samples from set-c.
- splits
- Includes 5 different splits of data indices (train, validation, test) to use them when running an algorithm five times to measure mean and standard deviation of the performance.
Raindrop was tested using Python 3.6 and 3.9.
To have consistent libraries and their versions, you can install needed dependencies for this project running the following command:
pip install -r requirements.txt
Text
Text
Raindrop is licensed under the MIT License.