| title | output | ||||
|---|---|---|---|---|---|
Assessing Virulence by MCG |
|
library("tidyverse")
dir.create(file.path(PROJHOME, "results", "tables"))This document is to assess virulence associated with the 10 most common MCGs. We
can get this from the cleaned data set that we saved in the
data-comparison.Rmd file. Note, we will be labelling the plot with
"Aggressiveness" as that is the preferred term.
datcols <- cols(
.default = col_integer(),
Severity = col_double(),
Region = col_character(),
Source = col_character(),
Host = col_character()
)
dat <- read_csv(file.path(PROJHOME, "data", "clean_data.csv"), col_types = datcols) %>%
select(Severity, MCG, Region, Source, Year)
dat## # A tibble: 366 x 5
## Severity MCG Region Source Year
## <dbl> <int> <chr> <chr> <int>
## 1 3.9 4 NE unk 2003
## 2 5.4 45 NE unk 2003
## 3 6.3 5 NY unk 2003
## 4 4.4 4 MN wmn 2003
## 5 4.7 4 MN wmn 2003
## 6 6.1 3 MI wmn 2003
## 7 5.5 5 MI wmn 2003
## 8 5.0 3 MI wmn 2003
## 9 5.2 3 MI wmn 2003
## 10 5.3 5 MI wmn 2003
## # ... with 356 more rows
Now, I can filter out the top 10 MCGs:
top_mcg <- dat %>%
group_by(MCG) %>%
summarize(N = n()) %>%
arrange(desc(N)) %>%
slice(1:10) %>%
inner_join(dat, by = "MCG") %>%
select(MCG, Severity) %>%
mutate(MCG = forcats::fct_inorder(as.character(MCG)))
top_mcg## # A tibble: 207 x 2
## MCG Severity
## <fctr> <dbl>
## 1 5 6.3
## 2 5 5.5
## 3 5 5.3
## 4 5 6.0
## 5 5 5.2
## 6 5 5.8
## 7 5 6.2
## 8 5 5.3
## 9 5 4.3
## 10 5 4.5
## # ... with 197 more rows
set.seed(2017-06-29)
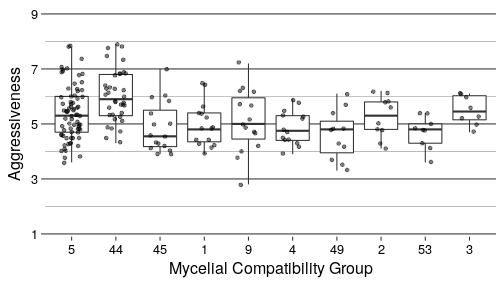
ggplot(top_mcg, aes(x = MCG, y = Severity)) +
geom_boxplot() +
geom_point(position = position_jitter(width = 0.25), alpha = 0.5) +
scale_y_continuous(limits = c(1, 9), breaks = c(1, 3, 5, 7, 9), expand = c(0, 0.1)) +
# scale_x_discrete(position = "top") +
theme_bw(base_size = 16, base_family = "Helvetica") +
theme(aspect.ratio = 1/2) +
theme(axis.text = element_text(color = "black")) +
# theme(axis.ticks.x = element_blank()) +
theme(panel.grid.major = element_line(colour = "grey20")) +
theme(panel.grid.minor = element_line(colour = "grey50")) +
theme(panel.grid.major.x = element_blank()) +
theme(panel.border = element_blank()) +
labs(list(
# title = "Aggressiveness for the top 10 MCGs",
y = "Aggressiveness",
x = "Mycelial Compatibility Group"
))top_mcg %>%
group_by(MCG) %>%
summarize(N = n(),
`Min Aggressiveness` = min(Severity),
`Max Aggressiveness` = max(Severity),
`Average Aggressiveness` = mean(Severity)
) %>%
knitr::kable(digits = 2)| MCG | N | Min Aggressiveness | Max Aggressiveness | Average Aggressiveness |
|---|---|---|---|---|
| 5 | 73 | 3.6 | 7.8 | 5.40 |
| 44 | 36 | 4.3 | 7.9 | 6.03 |
| 45 | 16 | 3.9 | 7.0 | 4.88 |
| 1 | 15 | 3.9 | 6.5 | 4.95 |
| 9 | 15 | 2.8 | 7.2 | 5.11 |
| 4 | 14 | 3.9 | 5.9 | 4.87 |
| 49 | 11 | 3.3 | 6.1 | 4.60 |
| 2 | 10 | 4.1 | 6.2 | 5.25 |
| 53 | 9 | 3.6 | 5.4 | 4.69 |
| 3 | 8 | 4.7 | 6.1 | 5.50 |
The default ANOVA in R sets contrasts as contrast.treatment, which compares
everything to the first factor, considered the treatment. Since we are
interested in whether or not there IS a difference between samples, this will
be sufficient.
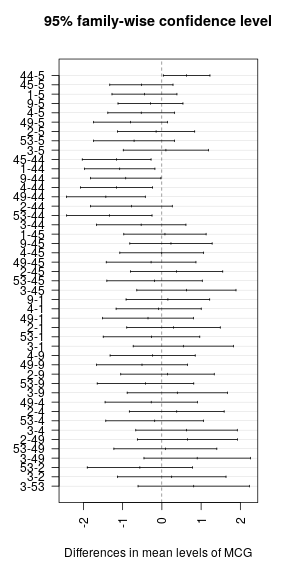
After the ANOVA, we performed a Tukey's Honest Significant Difference test to see exactly what groups these fell into.
ANOVA <- aov(Severity ~ MCG, data = top_mcg)
ANOVA## Call:
## aov(formula = Severity ~ MCG, data = top_mcg)
##
## Terms:
## MCG Residuals
## Sum of Squares 36.67876 163.83080
## Deg. of Freedom 9 197
##
## Residual standard error: 0.9119366
## Estimated effects may be unbalanced
summary(ANOVA)## Df Sum Sq Mean Sq F value Pr(>F)
## MCG 9 36.68 4.075 4.901 6.19e-06 ***
## Residuals 197 163.83 0.832
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
plot(TukeyHSD(ANOVA, conf.level = 0.95), las = 2)grps <- agricolae::HSD.test(ANOVA, "MCG", alpha = 0.05)$groups
grps %>%
tibble::rownames_to_column("MCG") %>%
dplyr::rename(Group = groups) %>%
dplyr::rename(`Mean Aggressiveness` = Severity) %>%
readr::write_csv(path = "results/tables/MCG-aggressiveness.csv", col_names = TRUE) %>%
huxtable::as_huxtable(add_colnames = TRUE) %>%
huxtable::set_number_format(row = huxtable::everywhere, col = 1, 0) %>%
huxtable::set_col_width(c(0.05, 0.3, 0.08)) %>%
huxtable::set_align(row = huxtable::everywhere, col = 2, "right") %>%
huxtable::print_md(max_width = 31)## ------------------------------
## MCG Mean Group
## Aggressiveness
## ---- ------------------ ------
## 44 6.03 a
##
## 3 5.50 ab
##
## 5 5.40 b
##
## 2 5.25 b
##
## 9 5.11 b
##
## 1 4.95 b
##
## 45 4.88 b
##
## 4 4.87 b
##
## 53 4.69 b
##
## 49 4.60 b
## ------------------------------
There appears to be a significant effect at p = 6.189e-06.
Since this was done in Otto-Hanson et al., it would be a good idea for us to assess this as well.
set.seed(2017-06-29)
region_dat <- dat %>%
group_by(Region) %>%
filter(n() > 5) %>%
mutate(mean_sev = mean(Severity)) %>%
mutate(Source = ifelse(Source == "wmn", "wmn", "producer field")) %>%
arrange(desc(mean_sev)) %>%
ungroup() %>%
mutate(Region = forcats::fct_inorder(Region)) %>%
select(Region, Severity, Source)
plot.mean <- function(x) {
m <- mean(x, na.rm = TRUE)
c(y = m, ymin = m, ymax = m)
}
set.seed(2017-09-06)
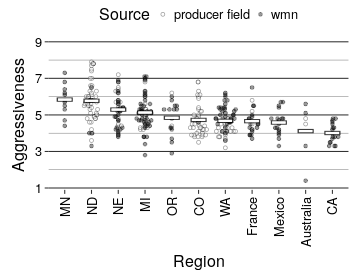
ggplot(region_dat, aes(x = Region, y = Severity)) +
ggforce::geom_sina(aes(fill = Source),
pch = 21,
binwidth = 0.1,
alpha = 0.4) +
# geom_point(aes(fill = Source),
# pch = 21,
# position = position_jitter(width = 0.25),
# alpha = 0.35) +
# geom_boxplot(alpha = 0.5) +
stat_summary(fun.data = plot.mean, geom = "errorbar", colour = "black", width = 0.6, size = 2, alpha = 0.5) +
stat_summary(fun.data = plot.mean, geom = "errorbar", colour = "white", width = 0.5, size = 1, alpha = 1) +
scale_y_continuous(limits = c(1, 9), breaks = c(1, 3, 5, 7, 9), expand = c(0, 0.1)) +
scale_fill_manual(values = rev(c("black", "white"))) +
theme_bw(base_size = 16, base_family = "Helvetica") +
theme(aspect.ratio = 1/2) +
theme(axis.text = element_text(color = "black")) +
theme(axis.text.x = element_text(angle = 90, hjust = 1, vjust = 0.5)) +
theme(panel.grid.major = element_line(colour = "grey20")) +
theme(panel.grid.minor = element_line(colour = "grey50")) +
theme(panel.grid.major.x = element_blank()) +
theme(panel.border = element_blank()) +
theme(legend.position = "top") +
theme(plot.margin = unit(c(0, 0, 0, 0), "lines")) +
labs(list(
y = "Aggressiveness",
x = "Region"
))if (!interactive()){
ggsave(file.path("results", "figures", "publication", "aggressiveness.pdf"),
width = 88, units = "mm")
}## Saving 88 x 95.2 mm image
region_dat %>%
group_by(Region) %>%
summarize(N = n(),
`Min Aggressiveness` = min(Severity),
`Max Aggressiveness` = max(Severity),
`Average Aggressiveness` = mean(Severity),
`Median Aggressiveness` = median(Severity)
) %>%
knitr::kable(digits = 2)| Region | N | Min Aggressiveness | Max Aggressiveness | Average Aggressiveness | Median Aggressiveness |
|---|---|---|---|---|---|
| MN | 11 | 4.4 | 7.3 | 5.84 | 5.80 |
| ND | 60 | 3.3 | 7.9 | 5.77 | 5.80 |
| NE | 47 | 3.8 | 7.2 | 5.29 | 5.30 |
| MI | 62 | 2.8 | 7.1 | 5.13 | 5.05 |
| OR | 17 | 2.9 | 6.2 | 4.84 | 5.30 |
| CO | 42 | 3.5 | 6.8 | 4.72 | 4.50 |
| WA | 59 | 3.2 | 6.2 | 4.67 | 4.60 |
| France | 22 | 3.7 | 6.5 | 4.66 | 4.50 |
| Mexico | 18 | 3.3 | 5.7 | 4.58 | 4.60 |
| Australia | 6 | 1.4 | 5.6 | 4.12 | 4.65 |
| CA | 18 | 3.3 | 4.8 | 4.01 | 3.90 |
ANOVA <- aov(Severity ~ Region, data = region_dat)
ANOVA## Call:
## aov(formula = Severity ~ Region, data = region_dat)
##
## Terms:
## Region Residuals
## Sum of Squares 86.07026 271.68215
## Deg. of Freedom 10 351
##
## Residual standard error: 0.8797859
## Estimated effects may be unbalanced
summary(ANOVA)## Df Sum Sq Mean Sq F value Pr(>F)
## Region 10 86.07 8.607 11.12 <2e-16 ***
## Residuals 351 271.68 0.774
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
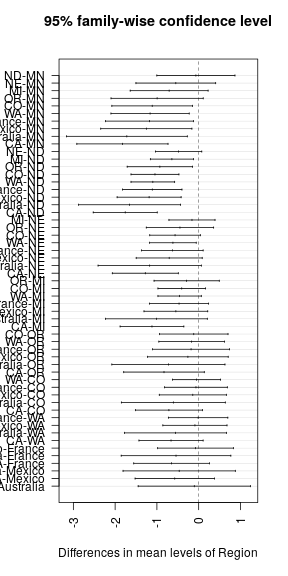
plot(TukeyHSD(ANOVA, conf.level = 0.95), las = 2)grps <- agricolae::HSD.test(ANOVA, "Region", alpha = 0.05)$groups %>%
tibble::rownames_to_column("Region") %>%
dplyr::rename(Group = groups) %>%
dplyr::rename(`Mean Aggressiveness` = Severity) %>%
readr::write_csv(path = "results/tables/Region-aggressiveness.csv", col_names = TRUE) %>%
huxtable::as_huxtable(add_colnames = TRUE) %>%
huxtable::set_number_format(row = huxtable::everywhere, col = 1, 0) %>%
huxtable::set_col_width(c(0.13, 0.25, 0.08)) %>%
huxtable::set_align(row = huxtable::everywhere, col = 2, "right") %>%
huxtable::print_md(max_width = 40)## -------------------------------------
## Region Mean Aggressiveness Group
## ---------- ------------------- ------
## MN 5.84 a
##
## ND 5.77 a
##
## NE 5.29 ab
##
## MI 5.13 abc
##
## OR 4.84 abcd
##
## CO 4.72 bcd
##
## WA 4.67 cd
##
## France 4.66 cd
##
## Mexico 4.58 cd
##
## Australia 4.12 cd
##
## CA 4.01 d
## -------------------------------------
The straw test, until the end of 2007, was performed by Lindsey Otto-Hanson. After that, these were performed by Serena McCoy. The Steadman lab was careful to train their members consistently in these practices, so the results from this test should be equivalent, but we want to ensure that there are no hidden biases between the two. To do this, we will test for differences within region.
set.seed(2017-06-29)
assessor_dat <- dat %>%
group_by(Region, Year) %>%
filter(n() > 5) %>%
mutate(mean_sev = mean(Severity)) %>%
mutate(Source = ifelse(Source == "wmn", "wmn", "producer field")) %>%
arrange(desc(mean_sev)) %>%
ungroup() %>%
mutate(Region = forcats::fct_inorder(Region)) %>%
mutate(Assessor = factor(ifelse(Year <= 2007, "Otto-Hanson", "McCoy"))) %>%
select(Region, Assessor, Severity, Source)
res <- aov(Severity ~ Region + Assessor, data = assessor_dat)
res## Call:
## aov(formula = Severity ~ Region + Assessor, data = assessor_dat)
##
## Terms:
## Region Assessor Residuals
## Sum of Squares 90.54166 0.29254 259.36272
## Deg. of Freedom 10 1 339
##
## Residual standard error: 0.8746895
## Estimated effects may be unbalanced
summary(res)## Df Sum Sq Mean Sq F value Pr(>F)
## Region 10 90.54 9.054 11.834 <2e-16 ***
## Assessor 1 0.29 0.293 0.382 0.537
## Residuals 339 259.36 0.765
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
library("poppr")## Loading required package: adegenet
## Loading required package: ade4
##
## /// adegenet 2.1.0 is loaded ////////////
##
## > overview: '?adegenet'
## > tutorials/doc/questions: 'adegenetWeb()'
## > bug reports/feature requests: adegenetIssues()
## This is poppr version 2.5.0. To get started, type package?poppr
## OMP parallel support: available
load(file.path("data", "sclerotinia_16_loci.rda"))
strat <- strata(dat11) %>%
add_column(MLG = mll(dat11)) %>%
add_column(Severity = other(dat11)$meta$Severity)
top_mlg <- strat %>%
group_by(MLG) %>%
summarize(N = n()) %>%
arrange(desc(N)) %>%
slice(1:10) %>%
inner_join(strat, by = "MLG") %>%
select(MLG, Severity) %>%
mutate(MLG = forcats::fct_inorder(as.character(MLG)))
set.seed(2017-06-29)
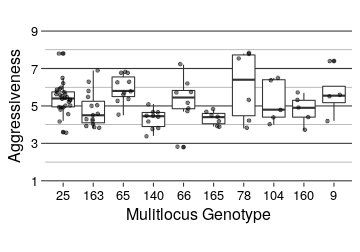
ggplot(top_mlg, aes(x = MLG, y = Severity)) +
geom_boxplot() +
geom_point(position = position_jitter(width = 0.25), alpha = 0.5) +
scale_y_continuous(limits = c(1, 9), breaks = c(1, 3, 5, 7, 9), expand = c(0, 0.1)) +
# scale_x_discrete(position = "top") +
theme_bw(base_size = 16, base_family = "Helvetica") +
theme(aspect.ratio = 1/2) +
theme(axis.text = element_text(color = "black")) +
# theme(axis.ticks.x = element_blank()) +
theme(panel.grid.major = element_line(colour = "grey20")) +
theme(panel.grid.minor = element_line(colour = "grey50")) +
theme(panel.grid.major.x = element_blank()) +
theme(panel.border = element_blank()) +
labs(list(
# title = "Aggressiveness for the top 10 MCGs",
y = "Aggressiveness",
x = "Mulitlocus Genotype"
))top_mlg %>%
group_by(MLG) %>%
summarize(N = n(),
`Min Aggressiveness` = min(Severity),
`Max Aggressiveness` = max(Severity),
`Average Aggressiveness` = mean(Severity)
) %>%
knitr::kable(digits = 2)| MLG | N | Min Aggressiveness | Max Aggressiveness | Average Aggressiveness |
|---|---|---|---|---|
| 25 | 27 | 3.6 | 7.8 | 5.41 |
| 163 | 15 | 3.8 | 6.9 | 4.80 |
| 65 | 11 | 4.5 | 6.8 | 5.94 |
| 140 | 10 | 3.4 | 5.1 | 4.31 |
| 66 | 8 | 2.8 | 7.2 | 5.30 |
| 165 | 7 | 3.9 | 4.8 | 4.34 |
| 78 | 6 | 3.8 | 7.8 | 6.07 |
| 104 | 5 | 4.0 | 6.5 | 5.22 |
| 160 | 5 | 3.7 | 5.7 | 4.80 |
| 9 | 4 | 4.2 | 7.4 | 5.67 |
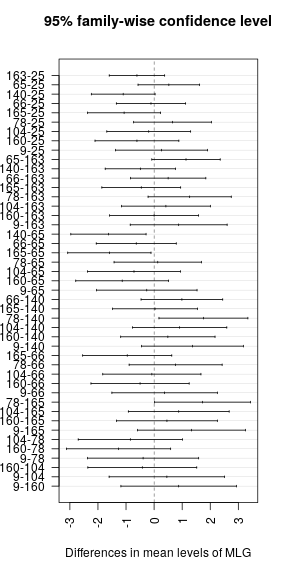
ANOVA <- aov(Severity ~ MLG, data = top_mlg)
ANOVA## Call:
## aov(formula = Severity ~ MLG, data = top_mlg)
##
## Terms:
## MLG Residuals
## Sum of Squares 28.91056 78.52710
## Deg. of Freedom 9 88
##
## Residual standard error: 0.9446446
## Estimated effects may be unbalanced
summary(ANOVA)## Df Sum Sq Mean Sq F value Pr(>F)
## MLG 9 28.91 3.212 3.6 0.000744 ***
## Residuals 88 78.53 0.892
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
plot(TukeyHSD(ANOVA, conf.level = 0.95), las = 2)grps <- agricolae::HSD.test(ANOVA, "MLG", alpha = 0.05)$groups %>%
tibble::rownames_to_column("MLG") %>%
dplyr::rename(Group = groups) %>%
dplyr::rename(`Mean Aggressiveness` = Severity) %>%
readr::write_csv(path = "results/tables/MLG-aggressiveness.csv", col_names = TRUE) %>%
huxtable::as_huxtable(add_colnames = TRUE) %>%
huxtable::set_number_format(row = huxtable::everywhere, col = 1, 0) %>%
huxtable::set_col_width(c(0.13, 0.25, 0.08)) %>%
huxtable::set_align(row = huxtable::everywhere, col = 2, "right") %>%
huxtable::print_md(max_width = 40)## ----------------------------------
## MLG Mean Aggressiveness Group
## ------- ------------------- ------
## 78 6.07 a
##
## 65 5.94 a
##
## 9 5.67 ab
##
## 25 5.41 ab
##
## 66 5.30 ab
##
## 104 5.22 ab
##
## 160 4.80 ab
##
## 163 4.80 ab
##
## 165 4.34 b
##
## 140 4.31 b
## ----------------------------------
Session Information
## Session info --------------------------------------------------------------------------------------
## setting value
## version R version 3.4.2 (2017-09-28)
## system x86_64, linux-gnu
## ui X11
## language (EN)
## collate en_US.UTF-8
## tz UTC
## date 2018-04-12
## Packages ------------------------------------------------------------------------------------------
## package * version date source
## ade4 * 1.7-8 2017-08-09 cran (@1.7-8)
## adegenet * 2.1.0 2017-10-12 cran (@2.1.0)
## agricolae 1.2-8 2017-09-12 cran (@1.2-8)
## AlgDesign 1.1-7.3 2014-10-15 cran (@1.1-7.3)
## ape 5.0 2017-10-30 cran (@5.0)
## assertr 2.0.2.2 2017-06-06 cran (@2.0.2.2)
## assertthat 0.2.0 2017-04-11 CRAN (R 3.4.2)
## base * 3.4.2 2018-03-01 local
## bindr 0.1 2016-11-13 CRAN (R 3.4.2)
## bindrcpp * 0.2 2017-06-17 CRAN (R 3.4.2)
## boot 1.3-20 2017-07-30 cran (@1.3-20)
## broom 0.4.3 2017-11-20 CRAN (R 3.4.2)
## cellranger 1.1.0 2016-07-27 CRAN (R 3.4.2)
## cli 1.0.0 2017-11-05 CRAN (R 3.4.2)
## cluster 2.0.6 2017-03-16 CRAN (R 3.4.2)
## coda 0.19-1 2016-12-08 cran (@0.19-1)
## colorspace 1.3-2 2016-12-14 CRAN (R 3.4.2)
## combinat 0.0-8 2012-10-29 cran (@0.0-8)
## compiler 3.4.2 2018-03-01 local
## crayon 1.3.4 2017-09-16 CRAN (R 3.4.2)
## datasets * 3.4.2 2018-03-01 local
## deldir 0.1-14 2017-04-22 cran (@0.1-14)
## devtools 1.13.4 2017-11-09 CRAN (R 3.4.2)
## digest 0.6.12 2017-01-27 CRAN (R 3.4.2)
## dplyr * 0.7.4 2017-09-28 CRAN (R 3.4.2)
## evaluate 0.10.1 2017-06-24 CRAN (R 3.4.2)
## expm 0.999-2 2017-03-29 cran (@0.999-2)
## ezknitr 0.6 2016-09-16 cran (@0.6)
## fastmatch 1.1-0 2017-01-28 cran (@1.1-0)
## forcats * 0.2.0 2017-01-23 CRAN (R 3.4.2)
## foreign 0.8-69 2017-06-21 CRAN (R 3.4.2)
## gdata 2.18.0 2017-06-06 cran (@2.18.0)
## ggforce 0.1.1 2016-11-28 cran (@0.1.1)
## ggplot2 * 2.2.1 2016-12-30 CRAN (R 3.4.2)
## glue 1.2.0 2017-10-29 CRAN (R 3.4.2)
## gmodels 2.16.2 2015-07-22 cran (@2.16.2)
## graphics * 3.4.2 2018-03-01 local
## grDevices * 3.4.2 2018-03-01 local
## grid 3.4.2 2018-03-01 local
## gtable 0.2.0 2016-02-26 CRAN (R 3.4.2)
## gtools 3.5.0 2015-05-29 cran (@3.5.0)
## haven 1.1.0 2017-07-09 CRAN (R 3.4.2)
## highr 0.6 2016-05-09 CRAN (R 3.4.2)
## hms 0.4.0 2017-11-23 CRAN (R 3.4.2)
## htmltools 0.3.6 2017-04-28 CRAN (R 3.4.2)
## httpuv 1.3.5 2017-07-04 CRAN (R 3.4.2)
## httr 1.3.1 2017-08-20 CRAN (R 3.4.2)
## huxtable 1.1.0 2017-10-20 cran (@1.1.0)
## igraph 1.1.2 2017-07-21 CRAN (R 3.4.2)
## jsonlite 1.5 2017-06-01 CRAN (R 3.4.2)
## klaR 0.6-12 2014-08-06 cran (@0.6-12)
## knitr * 1.17 2017-08-10 CRAN (R 3.4.2)
## lattice 0.20-35 2017-03-25 CRAN (R 3.4.2)
## lazyeval 0.2.1 2017-10-29 CRAN (R 3.4.2)
## LearnBayes 2.15 2014-05-29 cran (@2.15)
## lubridate 1.7.1 2017-11-03 CRAN (R 3.4.2)
## magrittr 1.5 2014-11-22 CRAN (R 3.4.2)
## MASS 7.3-47 2017-04-21 CRAN (R 3.4.2)
## Matrix 1.2-12 2017-11-16 CRAN (R 3.4.2)
## memoise 1.1.0 2017-04-21 CRAN (R 3.4.2)
## methods * 3.4.2 2018-03-01 local
## mgcv 1.8-22 2017-09-19 CRAN (R 3.4.2)
## mime 0.5 2016-07-07 CRAN (R 3.4.2)
## mnormt 1.5-5 2016-10-15 CRAN (R 3.4.2)
## modelr 0.1.1 2017-07-24 CRAN (R 3.4.2)
## munsell 0.4.3 2016-02-13 CRAN (R 3.4.2)
## nlme 3.1-131 2017-02-06 CRAN (R 3.4.2)
## parallel 3.4.2 2018-03-01 local
## pegas 0.10 2017-05-03 cran (@0.10)
## permute 0.9-4 2016-09-09 cran (@0.9-4)
## phangorn 2.3.1 2017-11-01 cran (@2.3.1)
## pkgconfig 2.0.1 2017-03-21 CRAN (R 3.4.2)
## plyr 1.8.4 2016-06-08 CRAN (R 3.4.2)
## poppr * 2.5.0 2017-09-11 cran (@2.5.0)
## psych 1.7.8 2017-09-09 CRAN (R 3.4.2)
## purrr * 0.2.4 2017-10-18 CRAN (R 3.4.2)
## quadprog 1.5-5 2013-04-17 cran (@1.5-5)
## R.methodsS3 1.7.1 2016-02-16 cran (@1.7.1)
## R.oo 1.21.0 2016-11-01 cran (@1.21.0)
## R.utils 2.6.0 2017-11-05 cran (@2.6.0)
## R6 2.2.2 2017-06-17 CRAN (R 3.4.2)
## Rcpp 0.12.14 2017-11-23 CRAN (R 3.4.2)
## readr * 1.1.1 2017-05-16 CRAN (R 3.4.2)
## readxl 1.0.0 2017-04-18 CRAN (R 3.4.2)
## reshape2 1.4.2 2016-10-22 CRAN (R 3.4.2)
## rlang 0.1.4 2017-11-05 CRAN (R 3.4.2)
## rstudioapi 0.7 2017-09-07 CRAN (R 3.4.2)
## rvest 0.3.2 2016-06-17 CRAN (R 3.4.2)
## scales 0.5.0 2017-08-24 CRAN (R 3.4.2)
## seqinr 3.4-5 2017-08-01 cran (@3.4-5)
## shiny 1.0.5 2017-08-23 CRAN (R 3.4.2)
## sp 1.2-5 2017-06-29 CRAN (R 3.4.2)
## spData 0.2.6.7 2017-11-28 cran (@0.2.6.7)
## spdep 0.7-4 2017-11-22 cran (@0.7-4)
## splines 3.4.2 2018-03-01 local
## stats * 3.4.2 2018-03-01 local
## stringi 1.1.6 2017-11-17 CRAN (R 3.4.2)
## stringr * 1.2.0 2017-02-18 CRAN (R 3.4.2)
## tibble * 1.3.4 2017-08-22 CRAN (R 3.4.2)
## tidyr * 0.7.2 2017-10-16 CRAN (R 3.4.2)
## tidyverse * 1.2.1 2017-11-14 CRAN (R 3.4.2)
## tools 3.4.2 2018-03-01 local
## tweenr 0.1.5 2016-10-10 cran (@0.1.5)
## udunits2 0.13 2016-11-17 cran (@0.13)
## units 0.4-6 2017-08-27 cran (@0.4-6)
## utils * 3.4.2 2018-03-01 local
## vegan 2.4-4 2017-08-24 cran (@2.4-4)
## withr 2.1.0 2017-11-01 CRAN (R 3.4.2)
## xml2 1.1.1 2017-01-24 CRAN (R 3.4.2)
## xtable 1.8-2 2016-02-05 CRAN (R 3.4.2)