Neural Spike Train Analysis. Allows one to import sorted Plexon files and perform a series of statistical analyses on neuronal discharge data through batch processing.
- Calculation Types
- SetUp Directories
- Prerequisite TODO
- Performing Calculations
- Visualization Functions
- Classification
- Calculations/Types of Analyses one can perform include:
- Measures of Central Tendency of ISI distribution: Standard deviation and Mean
- Mean FR
- CV
- CV2 (Nawrot et al., 2008)
- Approximate Entropy (Pincus 1991)
- dApEn (Darin, Soares, Wichmann 2006)
- Raster Plot Visualization
- Autocorrelogram Visualization
- 2-D ScatterPlot Visualization
- 3-D ScatterPlot Visualization
- Double Exponential (DExp) Fit of Autocorrelograms
- ISI Histogram Visualization
- ISI Converter
- Kurtosis
- Pearson's moment coefficient of Skewness
- Fano Factor (Darin, Soares, Wichmann 2006), (Eden & Kramer 2010)
- Non-parametric skew of ISI distribution
- Pearson mode Skew
- Spike Density Function
- Movement-specific Spike Density Function
- StatAv
- Coming Soon
- Hurst Exponent
Clone/download this repository and make sure to add all these folders to the current path in Matlab. Open_data.m, detecMove.m, and loadMoveFile are files that were created by a lab member Tim Whalen. I made edits to these files to make them compatible with the other files in this repository. These files are only included in this repo as they are needed to load the sorted neuron spike trains from Plexon.
 Select the "Add with Subfolders" option and add the directory that contains the .PL2 files you would like to examine.
Select the "Add with Subfolders" option and add the directory that contains the .PL2 files you would like to examine.
- Know how many classes of in-vivo data you would like to examine. This depends on the number of conditions you would like to study: Acute, Gradual, Alpha-Syn, etc. If you just want results for a given dataset, just use one condition.
- Make sure the condition you want is included in the switch cases of the dataInitializer.m file.

- Also include the condition in the master.m. Make sure to delete the condtions that are not being used from the typeNames array and the corresponding elseif statement. Add elseif statements for your condition and add the condition into the typeNames array.

-
Currently, there are eight cases (Acute, Alpha-Syn, Gradual, Gradual 35%, Gradual 65%, Naive, Unilateral Depleted, Unilateral Intact) in the dataInitializer.m file. It is important that you delete the cases along with the files and animalcodes. You will encounter an error unless you have the files listed in the dataInitializer.m file.
-
To add cases, follow this format:
case 'CONDITION_NAME' input.files = { %1 From animal one 'FILENAMEUNITONESorttt.pl2;' 'FILENAMEUNITTWOSorttt.pl2;' %2 From animal two 'FILENAMEUNITONESorttt.pl2;' 'FILENAMEUNITTWOSorttt.pl2;' 'FILENAMEUNITTHREESorttt.pl2;' }; input.animalcodes = [1 1 2 2 2]; data = open_data(input);
* **Please follow the naming convention of naming a PL2 file as 'fileNameAWsorttt-01.pl2'.**
* Keep in mind your animalcodes should correspond to the number of files you add.
- Create an excel file named 'custClassification.xlsx'. Create a separate sheet for each condition/case you have in dataInitializer.m.

Follow this format and define four columns that contain conditionFileName, conditionSPKC, conditionUnit, conditionRegularIrregularBurst'. The SpKC and unit information can be found via NeuroExplorer. Make sure to have a row in the excel sheet for each file you add in dataInitializer. If the class is unknown, simply put 'No Class'. Once you set the dataInitializer and custClassificaiton file up, you should be good to go.
The master Function will be the most important function you will use in batch processing a certain analysis script. The master function takes in three parameters:
- Handle of function you would like to apply (String). Functions are limited to functions in the Functions directory.
- The conditions you would like to apply (String). Conditions are 'FULL' (Applies to all conditions) , and the strings of the conditions you have defined: 'Acute', 'Alpha-Syn', etc.
- Third parameter: Will be updated in the future. Just use 0 for now.
%To calculate the statAv parameter for all conditions you have defined, put this command in the command window of Matlab
master('statAv', 'FULL', 0)
%To calculate the AppEntropy parameter for a condition 'condition_name' you have defined, put this command in the command window of Matlab:
master('AppEntropy', 'condition_name', 0)
This will create a results folder along with the results for each condition. Please refer to the function scripts to read about what they exactly calculate. The following functions can be used as function handles:
- 'AppEntropy'
- 'CV'
- 'CV2'
- 'ISIKurtosis'
- 'allstdISI'
- 'cellbycellISIAutos'
- 'dApEn' (only compatible once you calculate AppEntropy and shuffledApEn)
- 'classISIHists'
- 'spikedensityfunction'
- 'expFitAutos'
- 'expFitResults'
- 'fanoFactor'
- 'meanFR'
- 'meanISI'
- 'nonParametricSkew'
- 'pearsonModeSkew'
- 'pearsonSecSkew'
- 'sampleSkew'
- 'shuffledApEn'
- 'statAv'
- 'rasterPlot'
Parameters for these function handles can be adjusted in the mapper.m file.
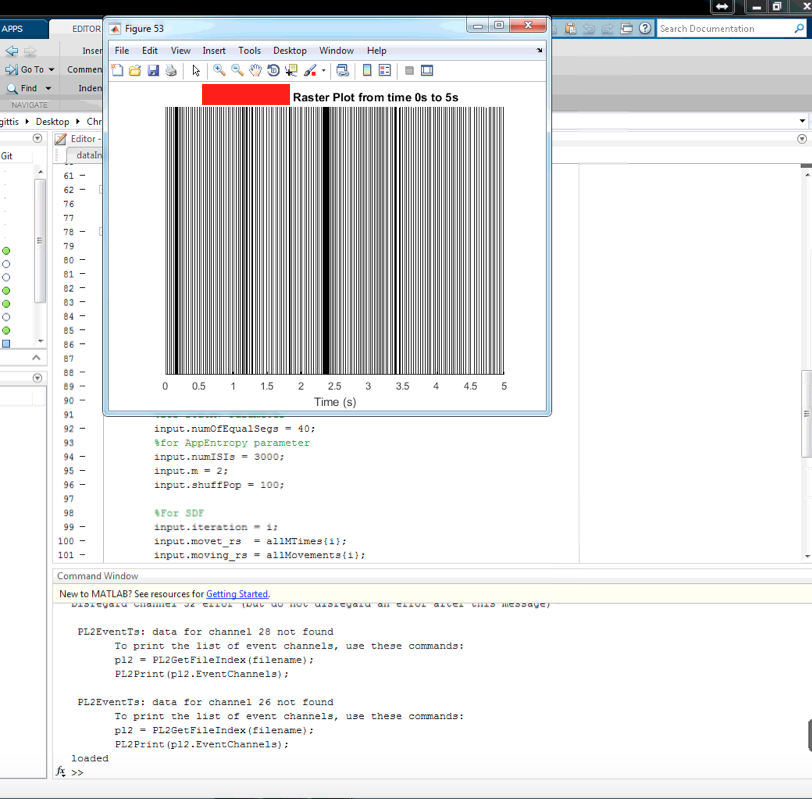
Rasterplot is a visualization function that can be used through the master function.
master('rasterPlot', 'condition_name', 0)
You should get a rasterPlot for each spike train in your file.
CellbycellISIAutos is a visualization script that can also be used through the master function.
master('cellbycellISIAutos', 'condition_name', 0)
Each row of a figure will have an ISI and autocorrelogram corresponding to a single spike train.
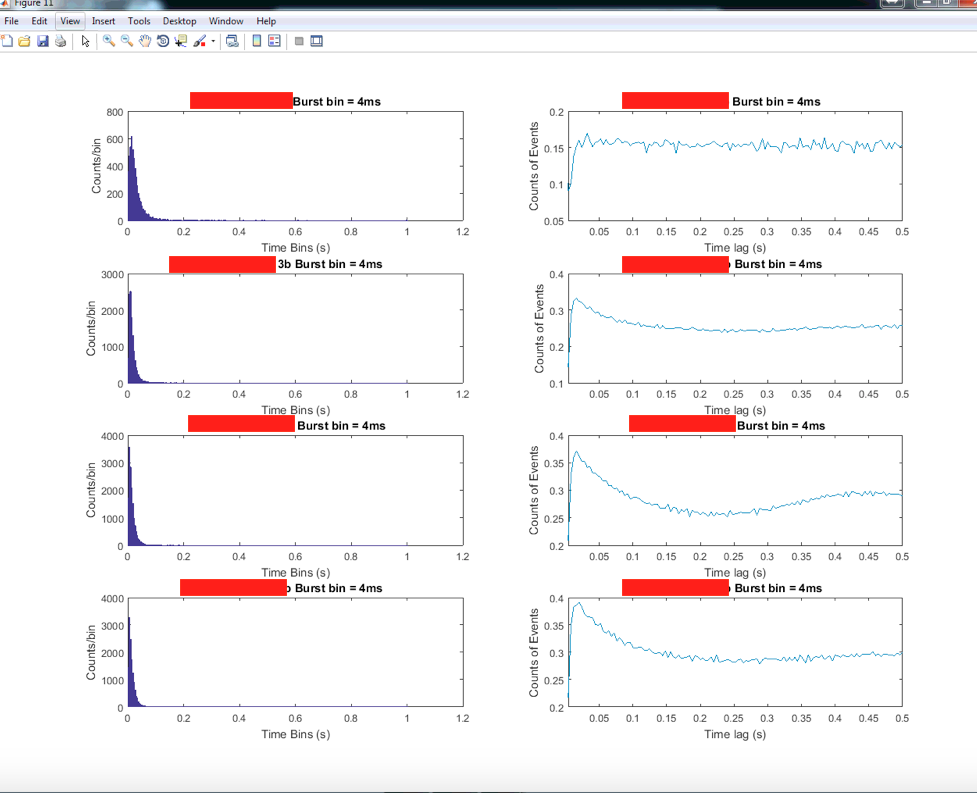
Empty subplots indicate that the given spike train did not fire above the threshold firing rate. The threshold firing rate can be changed in the cellbycellISIAutos.m script.
SpikeDensityFunction is a visualization script that can be used through the master function. It will show the spike density function results over multiple periods of movement.
You can adjust the time before movement onSet(line 31) and after movement onSet (line 32) in the spikedensityFunction.m file.
master('spikeDensityFunction', 'condition_name', 0)
 The first 3 subplots show that there are no movements/ lack of movement periods that fit the minimum time length. The last subplot shows different movement phases along with their spike density.
The first 3 subplots show that there are no movements/ lack of movement periods that fit the minimum time length. The last subplot shows different movement phases along with their spike density.
ScatterPlot allows you to produce a 2 dimensional scatterPlot that plots the results of one function against the results of another. Keep in mind, the ScatterPlot does NOT apply the functions to the datasets. Rather, it reads the results and then plots the data points in the 2 feature parameter space. Make sure to call the master function for the features first to get their results.
The scatterPlot function is not used via the master function. It takes in 3 input parameters:
- First Function handle (String)
- Second Function handle (String)
- Type of Conditions You'd like to examine (String) : 'FULL' creates scatterplots for ALL conditions.
Format of scatterPlot command:
scatterPlot('function_one', 'function_two', 'Type')
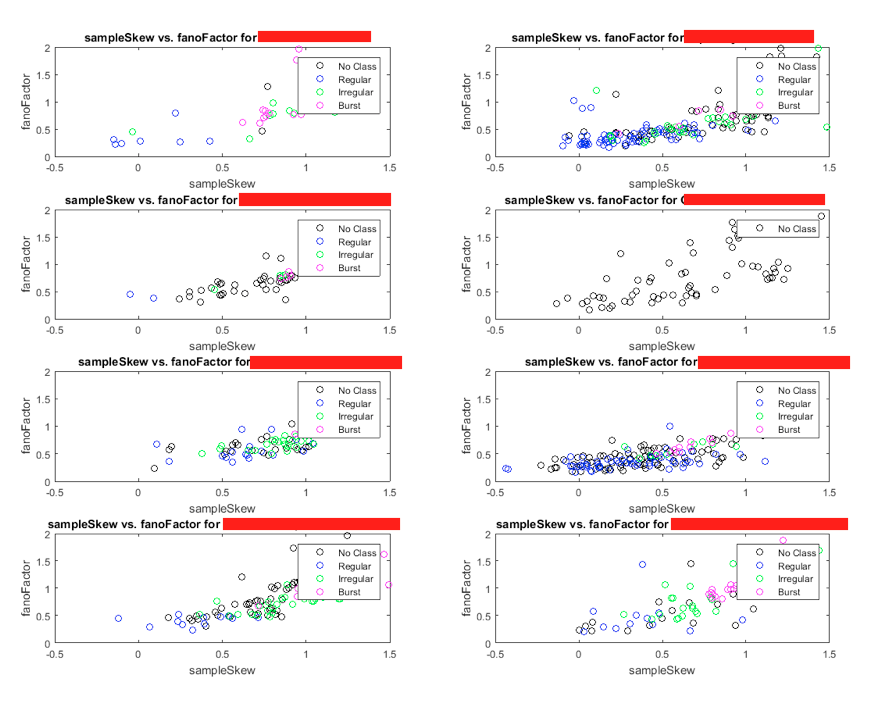
The plot above is produced when the statement
scatterPlot('sampleSkew', 'fanoFactor', 'FULL')
is put in the command window.
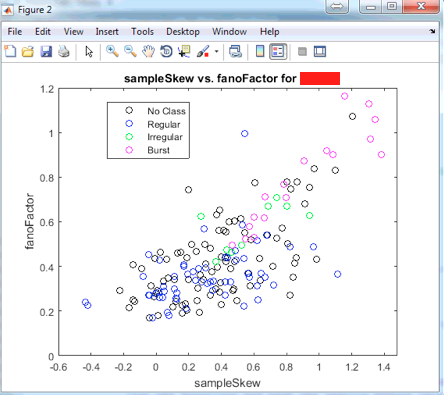
The plot above is produced when the statement
scatterPlot('sampleSkew', 'fanoFactor', 'Naive')
is put in the command window.
ScatterPlot3D allows you to produce a 3 dimensional scatterPlot that plots a series of data points in a 3D feature space. Keep in mind, the ScatterPlot3D does NOT apply the functions to the datasets. Rather, it reads the results and then plots the data points in the 3 feature parameter space. Make sure to call the master function for the features first to get their results.
The scatterPlot3d function is not used via the master function. It takes in 3 input parameters:
- First Function handle (String)
- Second Function handle (String)
- Third Function handle (String)
scatterPlot3d('function_one', 'function_two', 'function_three')
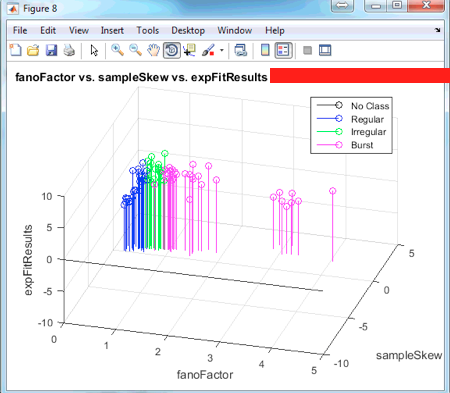
A figure like the one above is produced for EACH of the conditions if the command:
scatterPlot3d('fanoFactor', 'sampleSkew', 'expFitResults')
is used.

Press the button covered by the blue rectangle to rotate the 3D plot. Pressing the red rectangle will allow you to click on a certain data point and see its corresponding x,y,z values along with its fileName and SPKC unit.
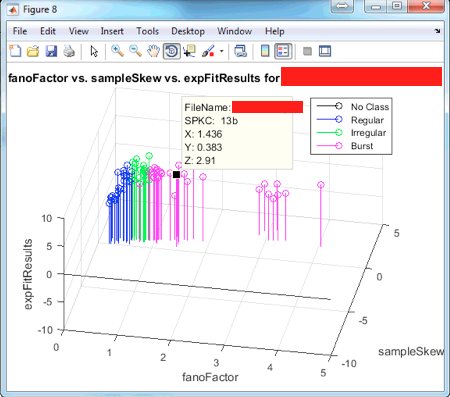
With the dataTip cursor, you can select a point and see something similar to what is shown above:
allscatterPlot3D plots all datapoints for all conditions in the SAME figure. Allscatterplot3d is useful in analyzing cumulative data (Similarities of features in Regular/Irregular/Burst neurons among ALL conditions).
This function takes 4 input parameters:
- First Function handle (String)
- Second Function handle (String)
- Third Function handle (String)
- Extra Functions cell (Cell)
The extra Functions cell should contain function handles that you'd like to see included in the dataTip. The extra Functions you use must also have their results already calculated (Must have used master function for extra functions). If you don't have any extra results you would like to see, just use an empty cell. AllScatterPlot3D also prints to the console the neurons classified as 'No Class'.
dataTipCell = {'extFunction1',..., 'extFunctionN'};
allscatterPlot3d('function_one', 'function_two', 'function_three', dataTipCell)

The following 3D plot is made when you type in the following commands:
dataTipCell = {'CV'};
allscatterPlot3d('sampleSkew', 'fanoFactor', 'expFitResults', dataTipCell)
This classification scheme employs the fanoFactor, sampleSkew, and doubleExpFit values to assign a neuron 1 of 4 possible types: No Class, Regular, Irregular, or Burst. IMPORTANT: To use this classification scheme, please make sure that you have calculated the results for the three parameters through the master function.
This function reads in the results of the three functions you want to use to reclassify the data. Based on the results of the three functions, the neurons will be classified by the criteria they meet. These threshold values can be adjusted in the reCustClassify.m script.
reCustClassify has three input parameters:
- First Function handle (String)
- Second Function handle (String)
- Third Function handle (String)
reCustClassify('function_one', 'function_two', 'function_three')
To use the classification scheme, you want to focus on the fanoFactor, sampleSkew, and expFitResults results. Thus, use this command:
reCustClassify('fanoFactor', 'sampleSkew', 'expFitResults')

This command will produce n (the number of conditions there are) condition_name results.csv files in your current directory. If you open any one of those files, they will have four columns: FileName, SPKC, Unit, Class. Import each of these files into the 'custClassification.xlsx' file. Each condition should have their own sheet in the excel file.
- Go to Data > From Text (In Get External Data Section) Select Delimited data type. Set the delimiters to be only commas when you import the .csv files. Press finish to import your newly classified data.

IMPORTANT Run the master function for all your functions again so that the classes for each neuron will be updated.
You may notice that some neurons are unclassified if your threshold values are not generalizable. You can fix this by creating broader threshold values or by manually inspecting the ISI histograms/Autocorrelograms of the 'No Class' neurons. Once you assign the class through visual inspection to each 'No Class' neuron, manually change the 'No Class' values in the 'custClassification.xlsx' to the values determined by visual inspection. Make sure to run the master function for all your functions again so that your classes will be updated.