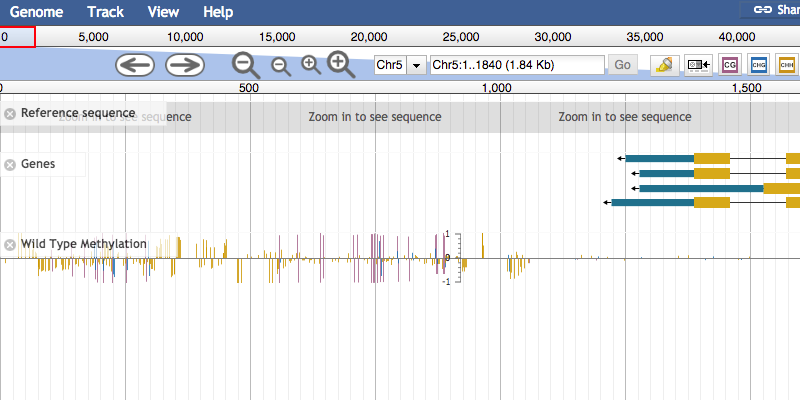
This is a JBrowse plugin
This plugin is to be used with whole-genome bisulfite sequencing data. Unlike almost all other browsers, this plugin allows you to see all methylation contexts on one track
For JBrowse 1.11.6+ in the JBrowse/plugins folder, type:
git clone https://github.com/bhofmei/jbplugin-methylation.git MethylationPlugin
or
downloaded the latest release version at releases.
Unzip the downloaded folder, place in JBrowse/plugins, and rename the folder MethylationPlugin
Add this to jbrowse.conf under [GENERAL]:
[ plugins.MethylationPlugin ]
location = plugins/MethylationPlugin
If that doesn't work, add this to jbrowse_conf.json:
"plugins" : {
"MethylationPlugin" : { "location" : "plugins/MethylationPlugin" }
}
DO NOT ADD THE PLUGIN TO BOTH!
NOTE: The plugin location folder can be named differently, i.e. jbplugin-methylation, but the plugin ID MUST be MethylationPlugin for the plugin to work correctly.
Sample data is included in the plugin to test that the plugin is working properly. With URL as the URL path to the JBrowse instance, navigate a web browser to URL/index.html?data=plugins/MethylationPlugin/test/data.
- Previous versions are supported for backwards compatability but using the current version (v3) is suggested.
- Version 1* is when all contexts are in a single BigWig file
- This caused problems with zooming in the browser, so version 2 was developed as a fix.
- Version 1 is still supported, but not recommended for use.
- Version 2 uses three BigWig files, one for each methylation context.
- The files need the same base name, like my-file.bw, and contexts are specified as additional extensions, i.e. my-file.bw.cg, my-file.bw.chg, and my-file.bw.chh.
- Visualization greatly depends on if the
-allparameter is used when converting the raw allC/bismark file
- Version 3 corrects the confusion about visualizating all sites vs methylated sites only
- Similar to version 2 in that there are 3 BigWig files (one for each context)
- Includes support to be able to filter use all or only methylated sites within one track. This keeps the global view looking good when zoomed out but allows for better visualization when zoomed in.
- Even for animals, who are interested in CG and CH, use the three contexts specified here. CHG and CHH are combined by the plugin to form CH context.
- Using the HTML-style track is not recommended because it can be slow for large regions and/or lots of data.
- However, HTML-style tracks are preferred when taking screenshots.
- The default methyl track type paints the basepair methylation (feature) on a HTML
canvaselement. - That's great for speed but is exported as an uneditable image.
- In the HTML-style tracks, each base (feature) is exported as an editable HTML
divelement
- The default methyl track type paints the basepair methylation (feature) on a HTML
- It is recommended you add all tracks as version 3 tracks and only use HTML-style tracks when taking screenshots. See this JBrowse plugin which makes taking screenshots easy and is configured to work with this methylation plugin.
- Note: The HTML-style track does allow for hiding/showing context and "methylated positions only" but does not support min and max score changes (default min: -1, max: 1).
The methylation tracks works best when the data is stored as BigWig file(s). File conversion is easy, though. Use the conversion program appropriate to your input file type. bedGraphToBigWig and bedSort (programs from UCSC) must be on your path. See details below for acquiring this program.
- allC files have these columns: chr, pos, strand, mc_class, mc_count, total, methylated
- In v2, by default, only methylated positions are included. Use the
-alloption to include all non-zero positions. - In v3, all non-zero positions are included. Methylated/unmethylated positions are denoted by the value.
- All chromosomes need to be combined into one allC file.
- You also need a file with chromosome sizes. This file should be tab-delimitated, one chromosome per line, with at least chromosome name and chromosome size. The genome's FASTA index (.fa.fai) file works well.
- For a given genome (i.e. chromosome sizes), you can specify an unlimited number of allC files to be converted.
- Note: version 1 conversion renames reference sequences to a convention while version 2 does not.
- Conversion Version 1:
Usage: python3 allc_old_bigwig_pe.py [-keep] [-no-clean] [-sort] [-all]
[-L=labels] [-p=num_proc] <chrm_sizes> <allC_file> [allC_file]*
-keep when on, keeps temporary files
-no-clean does not check chromosome names match chrm file
-sort calls bedSort; add this option if bigwig conversion fails
-all use all positions with methylation not just methylated ones
-L=labels comma-separated list of labels for incoming allC files
-p=num_proc number of processors to use [default 1]
-o=outID optional string to include in output file names
- Conversion Version 2:
Usage: python3 allc_to_bigwig_pe.py [-keep] [-sort] [-all] [-L=labels]
[-o=out_id] [-p=num_proc]<chrm_sizes> <allC_file> [allC_file]*
-keep keep intermediate files
-sort calls bedSort; add this option if bigwig conversion fails
-all use all positions with methylation [default methylated sites]
-allz use all positions with coverage including 0's
-L=labels comma-separated list of labels to use for the allC files;
defaults to using information from the allc file name
-o=out_id optional identifier to be added to the output file names
-p=num_proc number of processors to use [default 1]
- Conversion Version 3:
Usage: python3 allc_to_bigwig_pe_v3.py [-keep] [-sort] [-L=labels] [-p=num_proc]
[-o=out_id] <chrm_sizes> <allC_file> [allC_file]*
-keep keep intermediate files
-sort calls bedSort; add this option if bigwig conversion fails
-L=labels comma-separated list of labels to use for the allC files;
defaults to using information from the allc file name
-o=out_id optional identifier to be added to the output file names
-p=num_proc number of processors to use [default 1]
-
Bismark files have the following columns: chr, pos, strand, methylated reads, total reads, C context, trinucleotide context
-
There is a script to convert bismark files to version 2 and 3
-
Note: for v3, when filtering for methylated sites only, any site with at least 1 methylated read is considered "methylated"
-
Conversion Version 2
Usage: python3 bismark_to_bigwig_pe.py [-keep] [-sort] [-all] [-L=labels]
[-o=out_id] [-p=num_proc] <chrm_sizes> <bismark_file> [bismark_file]*
-keep keep intermediate files
-sort calls bedSort; add this option if bigwig conversion fails
-all use all covered positions including 0s [default only includes mC > 1]
-L=labels comma-separated list of labels to use for the allC files;
defaults to using information from the allc file name
-o=out_id optional identifier to be added to the output file names
-p=num_proc number of processors to use [default 1]
- Conversion Version 3
Usage: python3 bismark_to_bigwig_pe_v3.py [-keep] [-sort] [-L=labels] [-o=out_id]
[-p=num_proc] <chrm_sizes> <bismark_file> [bismark_file]*
-keep keep intermediate files
-sort calls bedSort; add this option if bigwig conversion fails
-L=labels comma-separated list of labels to use for the allC files;
defaults to using information from the allc file name
-o=out_id optional identifier to be added to the output file names
-p=num_proc number of processors to use [default 1]
Track specifications are very similar to those for XYPlots (see JBrowse tutorial for more information). The label, type, and urlTemplate must be specified.
{
"key" : "Wild Type Methylation",
"label" : "track_wild_type_methylation",
"style" : { "height" : 50 },
"storeClass" : "JBrowse/Store/SeqFeature/BigWig",
"urlTemplate" : "path/to/bigwig_file.bw",
"type" : "MethylationPlugin/View/Track/Wiggle/MethylXYPlot"
}
urlTemplate is the path and filename up-to, but not including, the context-specific extension. For example "urlTemplate" : "path/my-file.bw" from example above.
{
"key" : "Wild Type Methylation",
"label" : "track_wild_type_methylation",
"style" : { "height" : 50 },
"storeClass" : "MethylationPlugin/Store/SeqFeature/MethylBigWig",
"urlTemplate" : "path/to/bigwig_file.bw",
"type" : "MethylationPlugin/View/Track/Wiggle/MethylPlot"
}
If there is not a file for all contexts (CG, CHG, and CHH), include the following in the track configuration and specify only the contexts needed.
"contexts" : ["cg", "chh"]
Same as Version 2 except add this to configuration
"methylatedOption" : true
{
"type" : "MethylationPlugin/View/Track/MethylHTMLPlot",
"urlTemplate" : "line1-G3-rep1_Chr5_short.bw",
"category" : "Test data",
"maxHeight": 100,
"key" : "WT HTML Methylation",
"storeClass" : "MethylationPlugin/Store/SeqFeature/MethylBigWig",
"label" : "track_html_methylation"
}
While the plant world likes methylation broken into CG, CHG, and CHH, the animal world prefers CG and CH. For those only interested in CG, ignore this and be sure to specify only the CG context in the configuration (see above).
Using the animal coloring scheme is enforced hierarchically. Configurations specified at a higher level overpower lower-level specification. If not specified at a specific level, inherits the setting of the level below.
| level | location | syntax |
|---|---|---|
| highest | individual track config | isAnimal=true |
tracks.conf |
[general]isAnimal=true |
|
jbrowse.conf |
[general] isAnimal=true |
|
| lowest | default | isAnimal=false |
Note that toolbar buttons are defined by tracks.conf and jbrowse.conf.
- improved documentation for the different versions
Mac OSX 64-bit: https://hgdownload.cse.ucsc.edu/admin/exe/macOSX.x86_64/
Linux 64-bit: https://hgdownload.cse.ucsc.edu/admin/exe/linux.x86_64/
Older Linux/Linux server: https://hgdownload.cse.ucsc.edu/admin/exe/linux.x86_64.v287/
- Choose the appropriate web page from above. There will be a long list of programs.
- Scroll down to find bedGraphToBigWig and bedSort
- Save this program to computer
- In terminal, navigate to the directory with the program
- Type
chmod u+x bedGraphToBigWig bedSort - Move the program to the same directory as allc_to_bigwig_pe.py or add to path in .bashrc or .bash_profile
Versions of bedGraphToBigWig and bedSort are included in the bin directory.
Based on the operating system, create symbolic links in the bin directory
- MacOSX 64-bit:
ln -s bedGraphToBigWig_macOSX.x86_63 bedGraphToBigWig; ln -s bedSort_macOSX.x86_64 bedSort - Linux 64-bit:
ln -s bedGraphToBigWig_linux.x86_64 bedGraphToBigWig; ln -s bedSort_linux.x86_64 bedSort - Older Linux:
ln -s bedGraphToBigWig_linux.x86_64.v287 bedGraphToBigWig; ln -s bedSort_linux.x86_64.v287 bedSort
For best results, add this directory to PATH in .bashrc or .bash_profile.