Implemention of paper : Magicmol - A light-weighted pipeline for drug-like molecule evolution and quick chemical space exploration https://www.researchsquare.com/article/rs-2124702/v1
Magicmol provides a light-weighted pipeline for de novo novel molecule generation and evolution (combine with STONED-SELFIES), and can be utilized for either positive sampling or negative sampling. The process can be visulized in the following picture.
Original ChEMBL30 (https://www.ebi.ac.uk/chembl/) dataset, cleaned dataset are available at : https://drive.google.com/drive/folders/1ULI0ZxBk26EiCw_p0LnLB1ckgstWK0Hq?usp=sharing
190W: Original Chembl-30 small molecule dataset
database.cleaned.smi / database.smi : pure SMILES data / SMILES data cleaned by clean.py
database_smiles_0.25.pkl / database_smiles_0.5.pkl : random choosed 25% / 50% of the cleaned molecule data
python clean.py --in_path database.smi --out_path database_cleaned.smi --vocab_path ../vocab/chembl_selfies_vocab.yaml
The weight file is available at : https://drive.google.com/drive/folders/10LDOoQQzuL0XeDRdsWidhVsoN8W1wxFU?usp=sharing
Update: Model trained with SMILES is provided also in the above link.
Update: SMILES corpus and processing file are available now (see vocab).
However, we recommend you train the backbone model by yourself and leave it a cup of tea time (approximately 15 min on a single RTX3090) to finish this (XD) and experience how fast the process could be finished.
By default the training process is done by adopting SELFIES, more super parameters can be customized based on your requirements.
python train.py
For example, training with SMILES:
python train.py --num_embeddings 101 --vocab_path ./vocab/chembl_regex_vocab.yaml --which_vocab regex
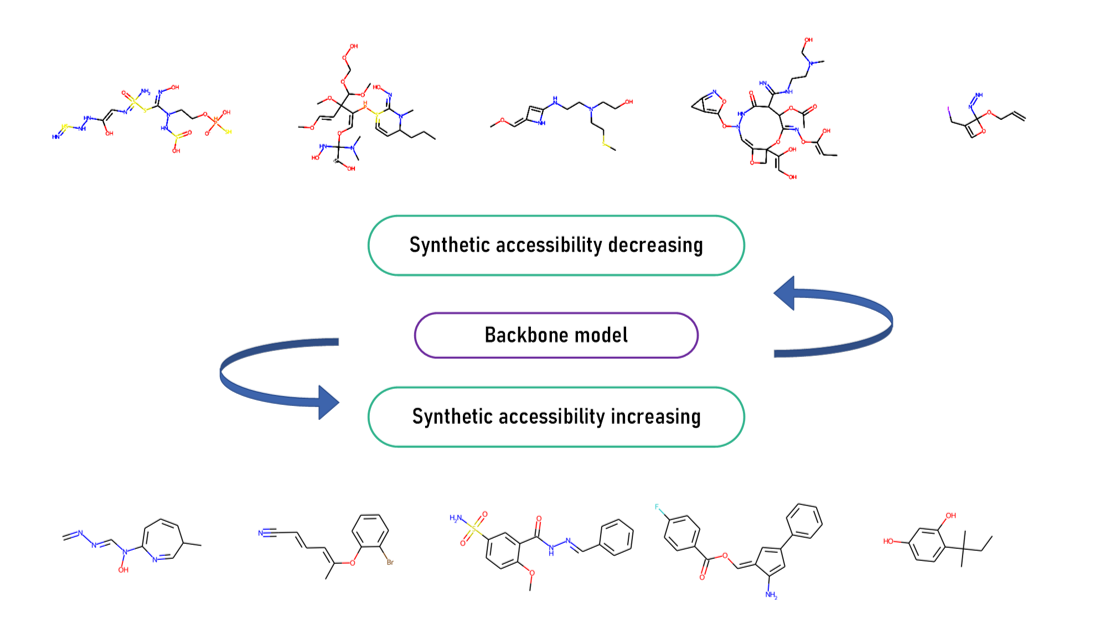
In our work, optimization is done by varying the SA score judged by SYBA from sampled molecules derived from the well-trained backbone model.
For positive optimization:
python synthetic_accessibility_modification.py --model_weight_path ../model_parameters/trained_model.pth --use_syba True --optim_task posi
For negative optimization:
python synthetic_accessibility_modification.py --model_weight_path ../model_parameters/trained_model.pth --use_syba True --optim_task nega
If you think our research is interesting, consider citing this paper? 👇
Lin Chen, Qing Shen, Jungang Lou et al. Magicmol - A light-weighted pipeline for drug-like molecule evolution and quick chemical space exploration, 10 October 2022, PREPRINT (Version 1) available at Research Square [https://doi.org/10.21203/rs.3.rs-2124702/v1]
STONED-SELFIES is available at: https://github.com/aspuru-guzik-group/stoned-selfies
SYBA is available at https://github.com/lich-uct/syba
Noted that our work was done by SELIFES 1.x, the above version may caused encoding problems.
Pickle needs python 3.8 or higher version for using pickle.HIGHEST_PROTOCOL.