Official PyTorch implementation of our method. The full paper is available at: Paper.
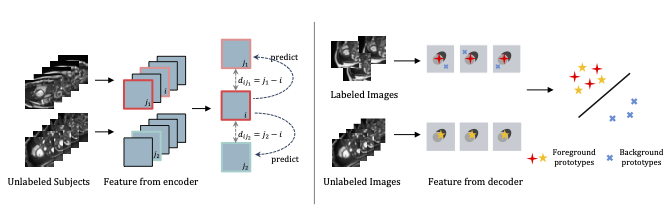
A novel bootstrap representation learning method by leveraging the predictable possibility of neighboring slices. At the core of our method are a simple and straightforward dense self-supervision on the predictions of local representations and a strategy of predicting locals based on global context, which enables stable and reliable supervision for both global and local representation mining among volumes.
# Preprocessing:
python preprocess.py
# Training:
python train.py --config=${CONFIG_NAME} --batch_size=${BATCH_SIZE} --seed=${RANDOM_SEED}
# Resume training:
python train.py --config=${CONFIG_NAME} --resume=${WEIGHTS_PATH} --batch_size=${BATCH_SIZE} --seed=${RANDOM_SEED}
# Testing:
python eval.py --config=${CONFIG_NAME} --seed=${RANDOM_SEED} --trained_model=best --no_sort --is_test --display
# Evaluation:
python evaluation.py --config=${CONFIG_NAME} --metric=${METRICS} --seeds=${RANDOM_SEEDS} --root=${EXP_FOLDER}You can create a definition in <configs/*_cfg.py>, then you can use any of the training commands in the previous section. (See the comments in <config.py> for an explanation of each field):
my_custom_config = Conifg({
'name': '',
'dataset': my_custom_dataset_config,
'transformer': my_custom_augmentation_config,
'model': my_custom_model_config,
'loss': my_custom_loss_config,
'optimizer': my_custom_optimizer_config,
# anything else.
})| Datasets | #Patients | DICE | Config Name |
|---|---|---|---|
| ACDC | 2 | 0.862 | pt_acdc_2p_config |
| ACDC | 8 | 0.899 | pt_acdc_8p_config |
| Prostate | 2 | 0.684 | pt_pst_2p_config |
| Prostate | 8 | 0.697 | pt_pst_8p_config |
| CAMUS_A2C | 8 | 0.813 | pt_a2c_8p_config |
| CAMUS_A2C | 32 | 0.868 | pt_a2c_32p_config |
| CAMUS_A4C | 8 | 0.832 | pt_a4c_8p_config |
| CAMUS_A4C | 32 | 0.878 | pt_a4c_32p_config |
if you find this code useful for your research, please cite:
@article{
title = {Bootstrap Representation Learning for Segmentation on Medical Volumes and Sequences},
author = {Zejian Chen and Wei Zhuo and Tianfu Wang and Wufeng Xue and Dong Ni},
journal = {CoRR},
volume = {abs/2106.12153},
year = {2021},
}
For questions about our paper or code, please contact Zejian Chen.