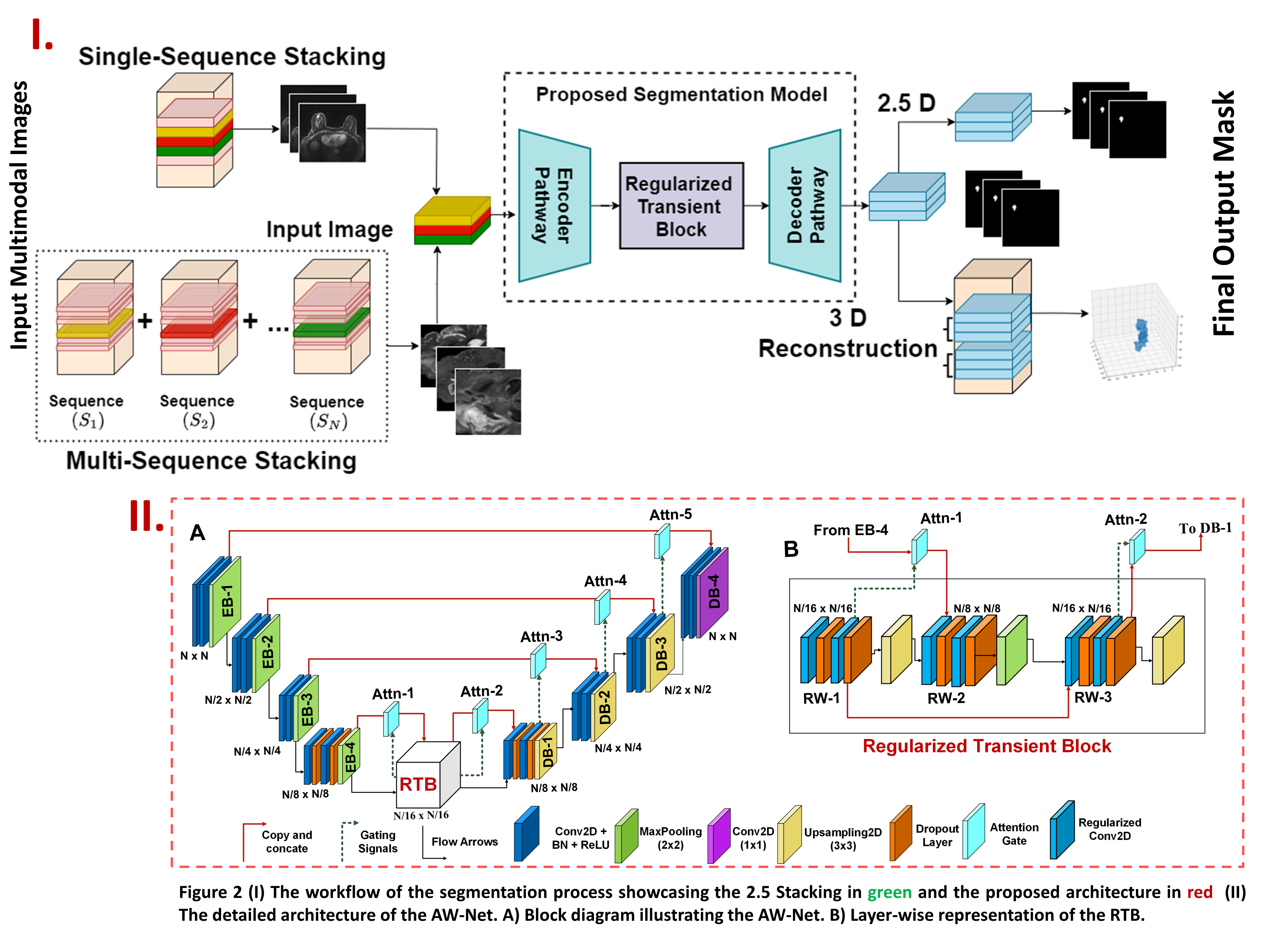
This repository contains the official codebase of AW-Net which was accepted in ICCV workshop 2023. AW-Net focuses on segmenting multimodal medical images such as MRI, PET, and CT scan images for multiple organs such as brain, breast, and spine.
A novel fully connected segmentation model which provides a solution to problem of segmenting multi-modal 3D/4D medical images by incorporating a novel regularized transient block. AW-Net uses L1 regularizers followed by dropout layers to improve model performance. The implementation is inspired from Attention UW-Net: A fully connected model for automatic segmentation and annotation of chest X-ray Code | Paper.
The datasets used in the paper can be downloaded from the links below:
The code is developed on tensorflow. In order to run the script file follow the instructions below:
- Create a new environment using
python -m /path/to/env - Fork the repository. Move the AW-Net folder to the new environment folder i.e. /path/to/env
- Use command
cd /path/to/env/AW-Netin the command prompt to access the files - There is a requirement.txt file. Run
pip install -r requirement.txtin the command prompt to install the required python packages.
We provide script files to train AW-Net on the BraTS2020 dataset. The dataloader.py works only for images and masks in numpy (.npy) format. The pre-processed version of BraTS2020 dataset can be accesed here link. The main.py contains script file to run the model. The following steps are required to execute the AW-Net architecture:
-
The function expects the path for the directory in which input images are present (arg1)
-
The function expects the path for the directory in which input masks are present (arg2)
-
The function expects the directory where the weights will be saved after the successful training of the model (arg3)
-
The default arguments in the function include num_classes (number of classes present in the mask), val_samples (number of images to be considered as a validation sample during splitting), epochs, and batch_size. Feel free to experiment on these as per the requirement.
-
In order to train the AW-Net model, run
cd /path/to/env/AW-Net python main.py arg1 arg2 arg3The saved weights will be stored under the path path/to/env/AW-Net/Data as .h5 file
In order to display the prediction generated by the model for a single image, run the following code snipet:
import numpy as np import matplotlib.pyplot as plt from model import* model=AW_Net((image_height,image_width,image_channel),num_classes, dropout_rate=0.0, batch_norm=True) model.compile(optimizer='adam',loss='categorical_crossentropy',metrics=['accuracy']) model.load_weights('path/to/env/AW-Net/Data/weights.h5') test_image = np.load('path/to/env/AW-Net/Data/Image/0.npy') predicted_image=model.predict(test_image.reshape(1,image_height,image_width,image_channel)) plt.imshow(np.argmax(predicted_image,axis=-1))
Set image_height = 128, image_width = 128, image_channel = 3, num_classes = 4 for BraTS2020 data.
If this code is used for research, please cite our paper:
Pal, Debojyoti, et al. "AW-Net: A Novel Fully Connected Attention-Based Medical Image Segmentation Model." Proceedings of the IEEE/CVF International
Conference on Computer Vision. 2023.
@inproceedings{pal2023aw,
title={AW-Net: A Novel Fully Connected Attention-Based Medical Image Segmentation Model},
author={Pal, Debojyoti and Meena, Tanushree and Mahapatra, Dwarikanath and Roy, Sudipta},
booktitle={Proceedings of the IEEE/CVF International Conference on Computer Vision},
pages={2532--2541},
year={2023}
}